DNA technology, the study and manipulation of genetic material has revolutionized modern science. Deoxyribonucleic acid (DNA) is a molecule that contains the biological instructions that make each species unique. DNA, along with the instructions it contains, is passed from adult organisms to their offspring during reproduction. Deoxyribonucleic acid (DNA), or an organism’s genetic material—inherited from one generation to the next—holds many clues that have unlocked some of the mysteries behind human behavior, disease, evolution, and aging.
The acronym “DNA” has become synonymous with solving crimes, testing for paternity, identifying human remains, and genetic testing. DNA can be retrieved from hair, blood, or saliva. Each person’s DNA sequences are unique, and it is possible to detect differences between individuals within a species on the basis of these unique features. DNA testing can also be used to identify pathogens, identify biological remains in archaeological digs, trace disease outbreaks, and study human migration patterns. In the medical field, DNA is used in diagnostics, new vaccine development, and cancer therapy. It is now also possible to determine predispositions to some diseases by looking at genes.
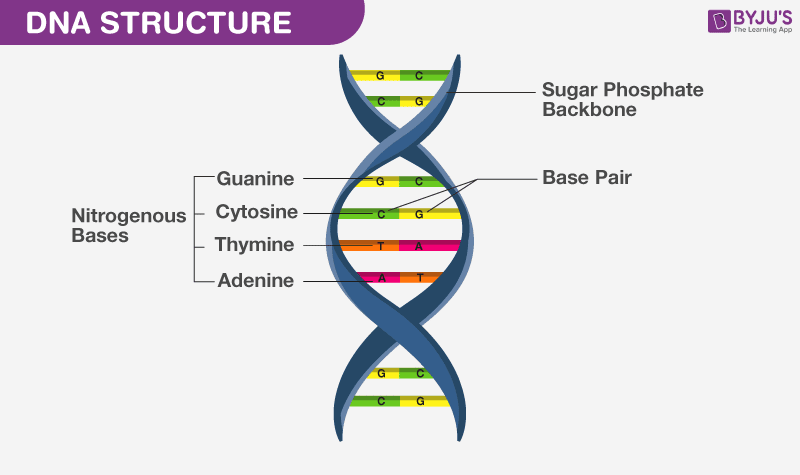
In organisms called eukaryotes, DNA is found inside a special area of the cell called the nucleus. Because the cell is very small, and because organisms have many DNA molecules per cell, each DNA molecule must be tightly packaged. This packaged form of the DNA is called a chromosome. Researchers refer to DNA found in the cell’s nucleus as nuclear DNA. An organism’s complete set of nuclear DNA is called its genome. DNA is made of chemical building blocks called nucleotides. These building blocks are made of three parts: a phosphate group, a sugar group and one of four types of nitrogen bases. To form a strand of DNA, nucleotides are linked into chains, with the phosphate and sugar groups alternating.
The four types of nitrogen bases found in nucleotides are: adenine (A), thymine (T), guanine (G) and cytosine (C). The order, or sequence, of these bases determines what biological instructions are contained in a strand of DNA. For example, the sequence ATCGTT might instruct for blue eyes, while ATCGCT might instruct for brown. The complete DNA instruction book, or genome, for a human contains about 3 billion bases and about 20,000 genes on 23 pairs of chromosomes.
Recent advances in DNA technology including cloning, PCR, recombinant DNA technology, DNA fingerprinting, gene therapy, DNA microarray technology, and DNA profiling have already begun to shape medicine, forensic sciences, environmental sciences, and national security.
Scientists are using DNA technology for a wide variety of purposes and products. A major component of DNA technology is cloning, which is the process of making multiple, identical copies of a gene. In cloning both the original organism and the clone have identical DNA. Identical twins are, in one sense, clones of each other; they have identical DNA, having developed from the same fertilized egg.
Cloning may bring to mind interesting sci-fi movies, but cloning also gives us pest-resistant plants, vaccines, heart attack treatments and even entirely new organisms. DNA technology has also had a major impact on the pharmaceutical industry, agriculture, disease therapy and even crime scene investigations.
PCR (polymerase chain reaction)
A novel tool called PCR (polymerase chain reaction) was developed that represents one of the most significant discoveries or inventions in DNA technology and it lead to a 1993 Nobel Prize award for American born Kary Mullis (1949–). PCR is the amplification of a specific sequence of DNA so that it can be analyzed by scientists.
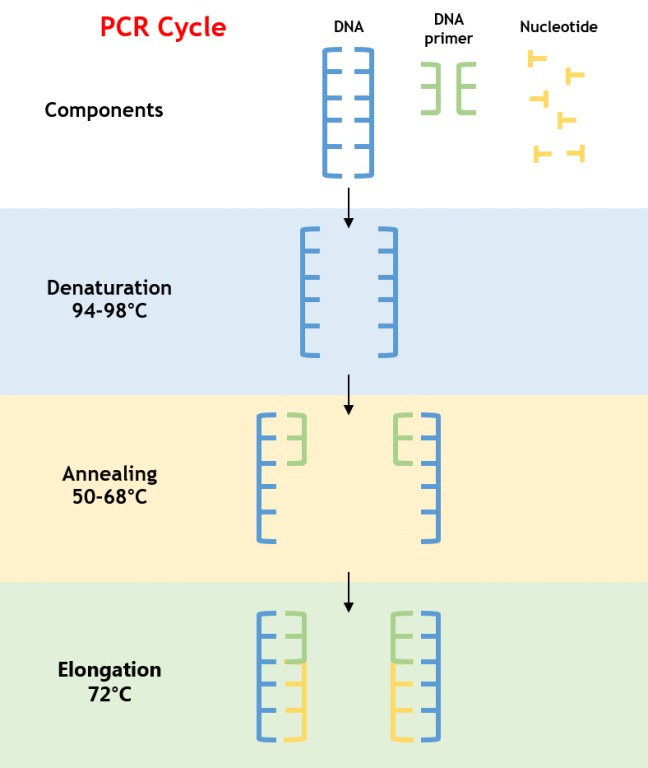
In traditional PCR, a strand of DNA is denatured, causing it to “unzip” and split into two halves. Next, a polymerase is added along with a DNA-coded primer designed for the genome of interest. The polymerase initializes a chemical process by which the two halves of the DNA strain are each “filled in” with the appropriate nucleic acid, creating two complete DNA molecules. This process is repeated, creating four molecules, then eight, then 16, and so on, exponentially increasing the total amount of DNA.
Amplification is important, particularly when it is necessary to analyze a small sequence of DNA in quantities that are large enough to perform other molecular analyses such as DNA sequencing.
Not long after PCR technology was developed, genetic engineering of DNA through recombinant DNA technology quickly became possible.
Recombinant DNA technology
Recombinant DNA is DNA that has been altered using bacterial derived enzymes called restriction endonucleases that act like scissors to cut DNA. The pattern that is cut can be matched to a pattern cut by the same enzymes from a different DNA sequence. The sticky ends that are created bind to each other and a DNA sequence can therefore be inserted into another DNA sequence.

Recombinant DNA technology can also be applied to splicing genes into molecular devices that can transport these genes to various cellular destinations. This technique, also called gene therapy, has been used to deliver corrected genes into individuals that have defective genes that cause disease. Gene splicing has also been applied to the environment as well.

The use of recombinant DNA technology has become commonplace as new products from genetically altered plants, animals, and microbes have become available for human use.Various bacteria have been genetically modified to produce proteins that break down harmful chemical contaminants such as DDT.
CRISPR (Clustered, Regularly-Interspaced Short Palindromic Repeats) allows scientists to edit genomes, far better than older techniques for gene splicing and editing. The CRISPR technique has enormous potential application, including altering the germline of humans, animals and other organisms, and modifying the genes of food crops. Ethical concerns have surfaced about this biotechnology and the prospect of editing the human germline and making so-called ‘designer babies’.
Creating a crop that is resistant to a specific herbicide proved to be a success because the herbicide eliminated weed competition from the crop plant. Researchers discovered herbicide-resistant bacteria, isolated the genes responsible for the condition, and “shot” them into a crop plant, which then proved to be resistant to that herbicide. Similarly, insect-resistant plants are becoming available as researchers discover bacterial enzymes that destroy or immobilize unwanted herbivores, and others that increase nitrogen fixation in the soil for use by plants. Currently, scientists are investigating the application of this technology to produce genetically engineered plants and crops that can produce substances that kill insects. Similarly, fruits can be engineered to have genes that produce proteins that slow the ripening process in an effort to extend their shelf life.
Medicine
Genetic engineering has resulted in a series of medical products. The first two commercially prepared products from recombinant DNA technology were insulin and human growth hormone, both of which were cultured in the E. coli bacteria.
Since then a plethora of products have appeared on the market, including the following abbreviated list, all made in E. coli:
- Tumor necrosis factor. Treatment for certain tumor cells
- Interleukin-2 (IL-2). Cancer treatment, immune deficiency, and HIV infection treatment
- Prourokinase. Treatment for heart attacks
- Taxol. Treatment for ovarian cancer
- Interferon. Treatment for cancer and viral infections
In addition, a number of vaccines are now commercially prepared from recombinant hosts. A vaccine is usually a harmless version of a bacterium or virus that is injected into an organism to activate the immune system to attack and destroy similar substances in the future. At one time vaccines were made by denaturing the disease and then injecting it into humans with the hope that it would activate their immune system to fight future intrusions by that invader. Unfortunately, the patient sometimes still ended up with the disease.
With DNA technology, only the identifiable outside shell of the microorganism is needed, copied, and injected into a harmless host to create the vaccine. This method is likely to be much safer because the actual disease-causing microbe is not transferred to the host. The immune system is activated by specific proteins on the surface of the microorganism -e. DNA technology takes that into account and only utilizes identifying surface features for the vaccine. Currently vaccines for the hepatitis B virus, herpes type 2 viruses, and malaria are in development for trial use in the near future.
Genetic fingerprinting
Genetic fingerprinting can be used to test for paternity. In forensics, genetic fingerprinting can be used to identify a criminal based on whether their unique DNA sequence matches to DNA extracted from a crime scene. This technology can also allow researchers to produce genetic maps of chromosomes based on these restriction enzyme fingerprints. Becasue there are many different enzymes, many different fingerprints can be ascertained.
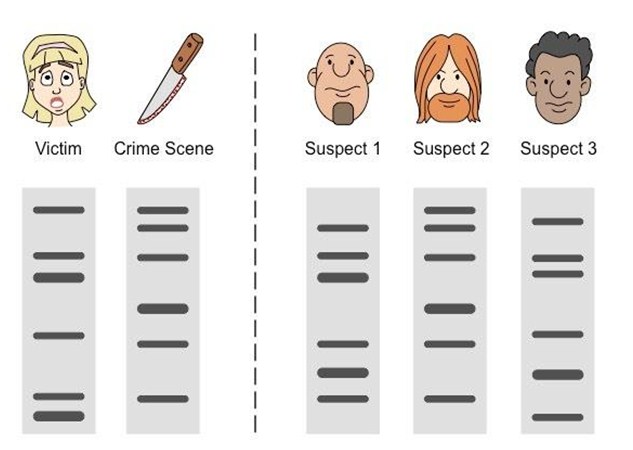
Colin Pitchfork Pitchfork was the first murderer to be caught using DNA analysis using the technique devised by Jeffrey Dahmer . Jeffreys had put his DNA pattern recognition technique to work in paternity and immigration cases, but now the police wanted him to help solve Ashworth’s murder. 1983. When Pitchfork’s DNA was analyzed, it matched the crime scene samples. Pitchfork was arrested and convicted and sentenced to life in prison. DNA profiling methods have become faster, more sensitive, and more user-friendly since the first murderer was caught with help from genetic evidence.
Although DNA evidence alone is not enough to secure a conviction today, DNA profiling has become the gold standard in forensic science since that first case 30 years ago. Despite being dogged by sample processing delays because of forensic lab backlogs, the technique has gotten progressively faster and more sensitive: Today, investigators can retrieve DNA profiles from skin cells left behind when a criminal merely touches a surface. This improved sensitivity combined with new data analysis approaches has made it possible for investigators to identify and distinguish multiple individuals from the DNA in a mixed sample. And it’s made possible efforts that are under way to develop user-friendly instruments that can run and analyze samples in less than two hours.
Chromosomes contain markers where short DNA sequences are repeated multiple times. The number of repeats at each marker varies from person to person, and each person has two copies, or alleles, of each marker, one inherited from their mother and one from their father. DNA contains regions in which short sequences of bases are repeated multiple times. These repeats are found in many spots—or loci—throughout the genome. Because the exact number of repeats at any particular locus varies from person to person, forensic scientists can use these markers, called short tandem repeats (STRs), to identify individuals. What’s more, people inherit chromosomes from both of their parents, so individuals have two sets of STRs, each of which can have different numbers of repeats at the same locus. This pair of alleles can therefore provide even more specificity to a person’s DNA profile.

During DNA profiling, cells are collected and broken open to gain access to their DNA. Then forensic scientists copy the DNA regions of interest and measure the length of the repeat sequences at multiple loci. The length rather than the exact sequence of the repeats serves as a marker for DNA profiles because repeat length is sufficient for distinguishing among individuals.
Probabilistic genotyping
Today’s forensic scientists are using mathematical methods that allow them to incorporate all the data in their analysis. Software packages use algorithms to determine which combinations of DNA profiles better explain the observed data. “It turns out, of all the trillion trillion or so possible explanations, most of them don’t really explain the data very well,” says Mark Perlin, chief executive and chief scientific officer of Cybergenetics, the producer of TrueAllele, which was the first major statistical software for analyzing complex DNA evidence.
This mathematical approach to DNA data interpretation is known as probabilistic genotyping. The software proposes genotypes for possible contributors to a DNA mixture and adds them together to construct datalike patterns. The software gives higher probability to proposed patterns that better fit the data. A Markov chain Monte Carlo algorithm ensures a thorough search and finds explanatory genotypes.
Belarusian, Russian scientists working on technology to portray criminals using DNA data
Belarusian and Russian scientists are working on a technology that will use DNA data to determine the ethnographic origin, age, and appearance of criminals in line with the DNA Identification program of the Union State of Belarus and Russia, BelTA learned from Irma Mosse, Head of the Human Genetics Lab of the Genetics and Cytology Institute of the National Academy of Sciences of Belarus (NASB), at a press conference on 20 February. The program began in 2017 and is designed to last for five years.
“Genetic technologies are widely used in healthcare and forensics. DNA samples can be collected from any biological material. Knowing the genotype of a person, we can tell a lot about him or her. These technologies are well developed for the sake of, for instance, identifying whether the DNA sample belongs to the suspect or not.
But science moves on. We have to learn how to determine what a person looks like – the color of eyes and hair, skin, height, body build, age, ethnographic origin. The psychoemotional state of the person is another important thing. These technologies are being developed as part of the Union State program DNA Identification.
We hope we will be able to portray a person using DNA samples by the time the work is done,” Irma Mosse said. In addition to finding criminals the DNA identification technology can help ascertain the identity of dead people or people, who have lost their memory. The Union State program also provides for developing innovative technologies for healthcare applications. In particular, plans have been made to study genetic predisposition to various diseases (cardiovascular, autoimmune, endocrine, and musculoskeletal ones).
There are plans to put together sets of reagents to find genetic markers to measure the risk of someone catching specific diseases. “We’ve started working out methods. We now have collections of samples. We’ve acquired the necessary equipment and reagents. The first results have been achieved with regard to the DNA identification of appearances, genetic determination of psychoemotional peculiarities of people, DNA diagnosis of liability to diseases.
We still have years of experiments to do,” added the head of the lab. Sergei Borovko, Deputy Chief of the Central Office for Forensic Examination of the Central Staff of the State Forensic Examination Committee of Belarus, told those present how genetic analysis is used in criminal investigations in Belarus. “Every year the committee performs about 15,000 DNA tests as part of criminal and civil cases, upon requests of citizens. The available equipment and methods as well as the experience accumulated over the course of more than 20 years allow us to get unique results not only as part of ongoing criminal investigations but also in criminal cases of the past. Many investigations into grave crimes have been left unfinished for decades.
The investigators have been able to finish them thanks to results of DNA tests. In 2017-2018 alone the DNA database grew larger with information about more than 150,000 genotypes of people, traces from sites of unresolved crimes, unidentified bodies. As the database grows, it becomes more useful for law enforcement agencies. In 2018 we sent about 2,000 messages about positive checks against the DNA database. Many of them helped solve crimes,” he noted.
Rapid DNA program
Today’s forensic scientist’s measure the size of DNA fragments with a technique called capillary electrophoresis. Small fragments travel more quickly than large fragments through a gel-like material. As the separated DNA bits pass a fluorescence detector, they are registered as a series of peaks in an electropherogram. Capillary electrophoresis instruments are much faster, and they don’t require users to cast gels as is done in the X-ray-based gel electrophoresis technique of the past, VCU’s Dawson Cruz says. “The system is much more automated than before.”
Not only has DNA analysis gotten faster it’s also breaking out of the lab. The U.S. government’s Rapid DNA program aims to develop integrated systems that work without human intervention so that nontechnical personnel could use them in police stations to query databases. The rapid DNA systems perform the same purification, amplification, separation, and detection steps that laboratories do. Software generates a profile that investigators can use to search the CODIS database for matches. Because the DNA from a reference sample comes from a single source, the system doesn’t need to perform mixture deconvolution.
Chinese authorities are collecting DNA — They say a comprehensive DNA database could be used to chase down any Uighurs who resist conforming to the campaign. It uses DNA to Track Its People. It wants to make the country’s Uighurs, a predominantly Muslim ethnic group, more subservient to the Communist Party. In Xinjiang, in northwestern China, the program was known as “Physicals for All.” From 2016 to 2017, nearly 36 million people took part in it, according to Xinhua, China’s official news agency. The authorities collected DNA samples, images of irises and other personal data, according to Uighurs and human rights groups. To bolster their DNA capabilities, scientists affiliated with China’s police used equipment made by Thermo Fisher, a Massachusetts company. For comparison with Uighur DNA, they also relied on genetic material from people around the world that was provided by Kenneth Kidd, a prominent Yale University geneticist.
DNA chip
DNA microarray technology, also known as the DNA chip, is the latest in nanotechnology that allows researchers the have ability to study the genome in a high throughput manner. It can be used for gene expression profiling which gives scientists insights into what genes are being up or down-regulated. Various genetic profiles can be determined in order to estimate cancer risk or to identify markers that may be associated with disease. It has the ability only to detect changes in gene expression that are large enough to be detected above a baseline level. Therefore, it does not detect subtle changes in gene expression that might cause disease or play a role in the development of disease. It can also be used for genotyping, although clinical diagnostic genotyping using microarray technology is still being investigated.
NetBio uses microfluidic chips for rapid DNA system
NetBio, is one of the companies that makes a rapid DNA system, also called Ande. Richard Selden, company founder and chief scientific officer, says the chemistry is the same as in conventional DNA analysis, but it’s much faster. The instrument decreased the time needed for PCR amplification from four hours to 17 minutes, he says.
Ande can be so much faster because it uses microfluidic chips and a very fast thermal cycler. With a typical PCR reaction, most of the time is spent ramping the temperature up and down. With the Ande system, “over the course of 15 minutes, you’re wasting a few minutes heating and cooling, but most of the time is the reaction being done,” Selden says.
The chip integrates all the steps of a typical DNA analysis. First, the cells are broken open and the DNA purified. Then the target loci are amplified. Finally, the amplified DNA is separated by electrophoresis and the sizes of the repeat segments determined. At the end of the process, about 90 minutes, the system automatically interprets the data to determine a profile, which is used to query CODIS or local DNA databases.
Synthetic DNA Market
Global Synthetic DNA Market is projected to reach US$4.2 Billion by 2028.
Synthetic DNA constructs are designed and manipulated using computer-aided design software. The designed DNA is then divided into synthesizable pieces (synthons) up to 1–1.5 kbp. The synthons are then broken up into overlapping single-stranded oligonucleotide sequences and chemically synthesized.
Synthetic biology is a field of science that involves redesigning organisms for useful purposes by engineering them to have new abilities. Synthetic biology researchers and companies around the world are harnessing the power of nature to solve problems in medicine, manufacturing and agriculture. DNA polymerase adds nucleotides to the deoxyribose (3′) ended strand in a 5′ to 3′ direction. Nucleotides cannot be added to the phosphate (5′) end because DNA polymerase can only add DNA nucleotides in a 5′ to 3′ direction. The lagging strand is therefore synthesised in fragments.
Synthetic DNA Geographical Segmentation Includes: – North America (U.S., Canada, Mexico) – Europe (U.K., France, Germany, Spain, Italy, Central & Eastern Europe, CIS); – Asia Pacific (China, Japan, South Korea, ASEAN, India, Rest of Asia Pacific); – Latin America (Brazil, Rest of L.A.); – Middle East and Africa (Turkey, GCC, Rest of Middle East)
Key players includes: Codexis, Inc., Creative Enzymes, Cyrus Biotechnology Inc., Eurofins Scientific, Integrated DNA Technologies, Inc.,
New England Biolabs, Synthego Corporation, Synthetic Genomics, Inc., TeselaGen, and Twist Bioscience
References and Resources also include:
https://science.jrank.org/pages/2134/DNA-Technology.html
 International Defense Security & Technology Your trusted Source for News, Research and Analysis
International Defense Security & Technology Your trusted Source for News, Research and Analysis
