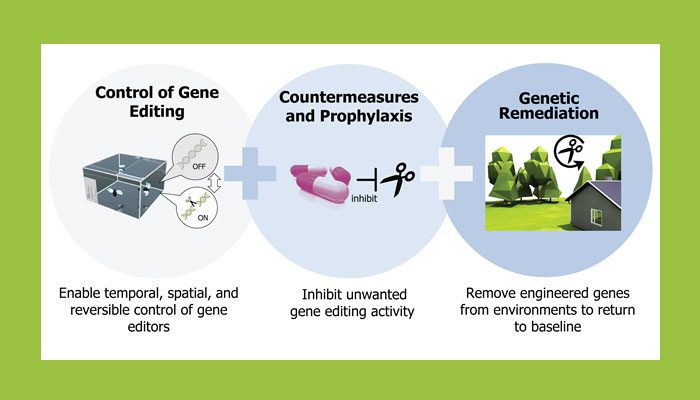
Gene editing technologies have captured increasing attention from healthcare professionals, policymakers, and community leaders in recent years for their potential to selectively disable cancerous cells in the body, control populations of disease-spreading mosquitos, and defend native flora and fauna against invasive species, among other uses. The potential national security applications and implications of these technologies are equally profound, including protection of troops against infectious disease, mitigation of threats posed by irresponsible or nefarious use of biological technologies, and enhanced development of new resources derived from synthetic biology, such as novel chemicals, materials, and coatings with useful, unique properties, says DARPA.
IDST Pro Access Required
This analysis is part of IDST premium intelligence.
Subscribe to Continue ReadingAlready a member? Log in